Chemotaxis
Bacterial chemotaxis is the mechanism by which bacteria sense and move in response to chemical signals in their environments; it is crucial in the virulence of pathogenic bacteria, and is linked to the formation of biofilms. A great deal is known about the various aspects of the bacterial chemosensory pathway, and the robust two-component signalling network of E.coli chemotaxis is one of the best understood signalling pathways in biology.
A decrease in the concentration of an attractant in the environment decreases the number of molecules of that attractant that are bound to specific receptors in the E. coli cell membrane. This signal is transmitted across the cell membrane to the cytoplasmic histidine protein kinase, CheA. The kinase activity of CheA is increased, and as a result there is more phosphorylation of the response regulator CheY. CheY-P binds to the flagellar motor and causes the motor to reverse its direction which results in the E. coli cell tumbling and changing direction. There is also an adaptation pathway consisting of CheR and CheB which resets the signalling state of the CheA to the prestimulus level so that the cell can sense subsequent changes in attractant concentration.
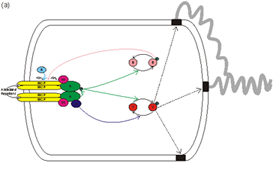
The structures, copy numbers, localisation, kinetics and dynamics of all of the main components of this system have been elucidated. As have the input:output characteristics and the effects of the deletion of most of the genes encoding these proteins. The wealth of existing data has made the E. coli chemotaxis system a prime target for systems biology and modelling approaches.
Whilst based on the simple pathway that is found in E. coli, the majority of bacterial chemotaxis signalling networks are vastly more complex. In general they possess multiple homologues of the components of the E. coli pathway along with a few proteins which are not used by E. coli. Our aim is to model and understand the relatively simple chemotaxis pathway in E. coli and then to extend this into our more complex model organism, Rhodobacter sphaeroides. This work also allows us to address the fundamental issue of scalability in the computational modelling of biological systems and in Systems Biology.
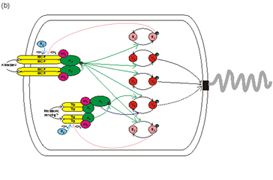
We are using a variety of different theoretical approaches to generate models of the chemotaxis pathways in E. coli and R. sphaeroides. These models then lead to predictions which can be tested experimentally and the results of the experiments fed back to the theoreticians in order to refine or refute different models. It is this strong iteration between theory and experiment that is essential for the success of modelling of this system and for all Systems Biology.
Mathematical Approaches to Systems Biology Research - Alex Fletcher
Multiscale Modelling of Key Components of the Chemosensing Pathway - Ben Hall
Localisation of Proteins in Chemotaxis Pathways - Rebecca Hamer & Pao-Yang Chen
Response kinetics of the Rhodobacter sphaeroides chemosensory pathway - Mila Kojadinovic
Biological Dynamics and Time Series Analysis for Systems Biology - Max Little, Sam Johnson
Computational Approaches to Systems Biology Research - James Osborne
Annotation of the R.sphaeroides WS8 genome and development of transcriptomic tools - Steven Porter, David Wilkinson
Multiscale Simulations and Experiments for Systems Biology - Kathryn Scott
Middleware and applications developer - Mark Slaymaker
Modelling Biological Networks - David Smith
Quantitative (Sub) Cellular Image Analysis - Quan Xue
Development and analysis of covariance based tools for determining protein:protein interaction specificity determining residues from protein sequences - Sonja Pawelczyk & Rebecca Hamer
Armitage
Bray
Gavaghan
Hein
Jones
Leake
Maini
Reinert
Sansom
Wadhams


